Forensic genetic genealogy operates in a fundamentally different way from consumer or academic pedigree analysis. Contemporary cases frequently involve imperfect SNP profiles, hundreds or thousands of individuals, incomplete or conflicting records, endogamy, misattributed parentage, and distant genetic relationships in which evidentiary value emerges only in combination. Despite these challenges, many practitioners continue to use analytical approaches that are rooted in assumptions that hold only for high quality, complete SNP profiles and small, well-resolved family trees.
Understanding why these approaches fail at scale requires reframing how forensic genetic genealogical inference is modeled.
A Positioning-Based Model of Genetic Genealogical Inference
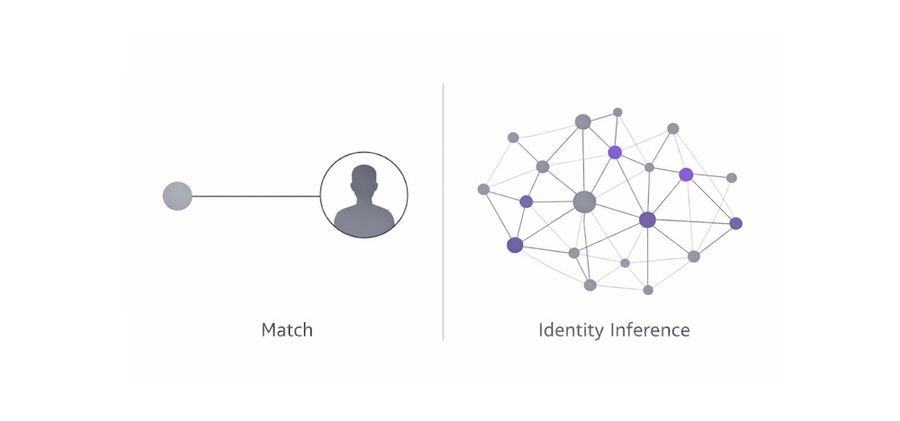
Genetic genealogical inference is essentially a problem of locating an unknown individual within a constrained relational structure, where position is defined by ancestry, descent, and inheritance rather than by pre-enumerated hypotheses.
The Genetic Genealogical Positioning System (GGPS), developed by the Othram Research team, models forensic genetic genealogy as a positioning problem over a genealogical graph, rather than as a sequence of manually defined hypotheses over a flat, fixed pedigree.
Instead of evaluating whether an unknown individual fits within a user-selected ancestral structure by effectively asking, “Does this person fit under this MRCA?”, GGPS frames the question from a different, more versatile perspective:
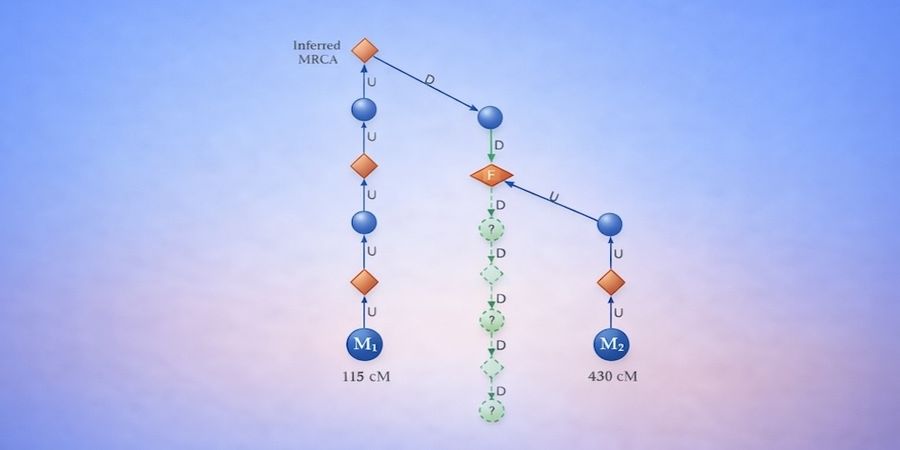
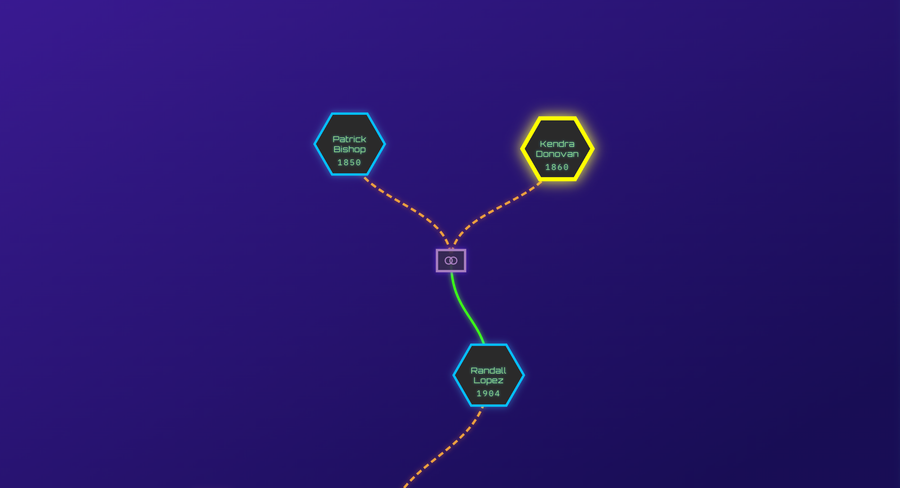
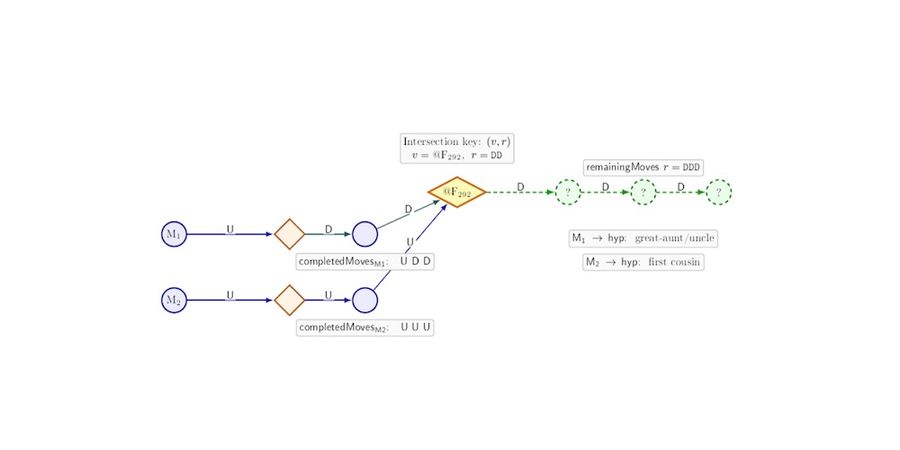
GGPS represents individuals and family units as nodes in a graph and systematically traverses biologically plausible genealogical paths upward and downward from known DNA associations. For each association, all compatible relationship paths supported by the shared DNA evidence are enumerated. The system then identifies convergence points where multiple independent paths intersect at the same location in the graph with the same remaining genealogical steps.
Each convergence thus defines a concrete and testable hypothesis about the placement of the unknown individual. Hypotheses are ranked using a unified score that balances combined DNA evidence, expected genealogical workload, and the likelihood of successfully resolving missing links.
In this framework, most recent common ancestors (MRCAs) are not inputs. They are outputs that emerge as structural consequences of path convergence when multiple independent DNA paths intersect within the graph. This shift in genealogy processing allows inference to proceed without requiring the practitioner to guess where in the tree the solution should lie before sufficient evidence has accumulated.
The Structural Limitations of MRCA-Anchored Tools
MRCA-anchored approaches require the analyst to pre-select the most recent common ancestor and restrict hypothesis evaluation to descendants of that ancestral couple. While this is tractable in small or well-curated pedigrees, it becomes increasingly fragile as genealogical complexity increases.
As family networks grow, the number of plausible MRCAs expands rapidly. Exhaustive manual evaluation becomes infeasible, forcing early pruning of the search space. Valid hypotheses are often excluded not because they are inconsistent with the data, but because they lie outside the initially chosen anchor.
These approaches also implicitly assume that the underlying tree structure is largely correct. In forensic contexts, this assumption is routinely violated due to record gaps, non-paternity events, adoption, and duplicated or conflated individuals. Anchoring inference to an incorrect MRCA amplifies structural error and obscures alternative explanations elsewhere in the graph.
Most critically, MRCA-anchored tools are not designed to discover convergence across multiple independent DNA associations. Convergence is evaluated only after an MRCA has been selected, rather than being used as the mechanism by which candidate ancestral structures are revealed. As the number of associations increases, this limitation becomes more pronounced.
At scale, MRCA-anchored tools invert the inference process by requiring structural commitment before evidentiary convergence has been established. Thus, quality investigative leads may be missed.
A Graph-Driven Alternative
GGPS was developed to address these limitations by operating directly on a genealogical graph rather than within a predefined hierarchical subtree. The system searches across the full connected structure, identifies where independent DNA-supported paths intersect, and surfaces hypotheses grounded in graph topology rather than user-selected anchors.
This approach allows inference to proceed in the presence of ambiguity, incompleteness, and local structural error. It also enables prioritization of hypotheses based on both evidentiary support and investigative tractability.
Data-driven positioning, instead of traditional anchored guessing, allows approaches to scale.
Advanced Analysis, Purpose-Built for Forensics
Two additional design decisions reflect the realities of working with forensic genetic genealogy data.
First, GGPS does not require all DNA associations to be consistent with a single hypothesis. Associations that are incompatible with a coherent solution, due to tree errors or missing information, are excluded from that hypothesis rather than forcing failure of the entire analysis.
Second, genealogical path selection incorporates X chromosome-DNA inheritance constraints. Plausible autosomal paths that violate X chromosome-DNA transmission rules are excluded at the path level, further constraining the hypothesis space and reducing biologically invalid placements.
From Anchored Hypotheses to Systematic Discovery
MRCA-anchored models are effective when genealogical structure is well understood. Forensic genetic genealogy rarely operates under such conditions. As case complexity increases, scalable inference requires moving from anchored hypothesis testing to systematic discovery across large and uncertain family networks.
GGPS represents one such positioning-based approach, grounded in graph traversal and evidentiary convergence, and is designed to operate under the structural realities of forensic genetic genealogy.
Our team is excited to make GGPS available to everyone, right now, within Othram Maps. Give it a try today at maps.othram.com.