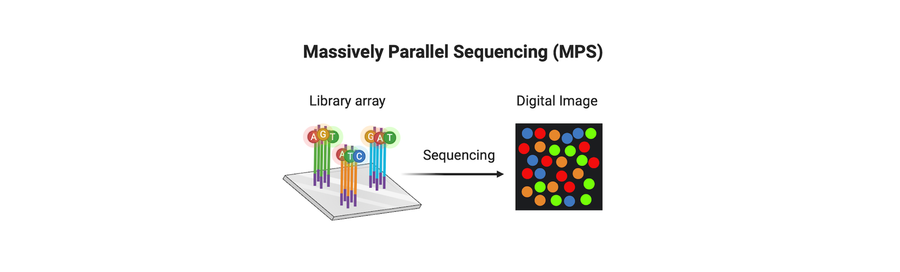
Forensic DNA testing has always required results that are defensible.
From its earliest use in criminal investigations and human identification, forensic genetics has operated under a higher standard than many other scientific disciplines. Results must be traceable, reliable as demonstrated through validation, peer review and publication, and capable of withstanding scrutiny in court. The question has never been whether or not a result could be generated, but whether or not the interpretations can be trusted, explained, and defended.
As forensic DNA analysis has evolved, the underlying technologies have changed, but these requirements remain constant.
The transition from short tandem repeat (STR) analysis to massively parallel sequencing (MPS) represents one of the most significant shifts in forensic genetics over the past two decades. MPS enables the generation of orders of magnitude more data from a single sample. It allows analysis of highly degraded DNA, supports genome-scale SNP profiling, and powers applications such as distant kinship inference, biogeographical ancestry analysis, and other advanced investigative tools.
With this shift comes a corresponding need for quality assurance (QA) and quality control (QC) systems that keep pace, not only in sensitivity but in auditability and defensibility.
Traditional forensic QA/QC frameworks were designed around targeted assays and relatively limited data output. MPS alters the simplicity of that equation. The same technologies that increase sensitivity and resolution also increase the complexity of laboratory workflows and the potential impact of error.
At the same time, MPS presents an unprecedented opportunity. The volume and richness of sequencing data make it possible to embed far more information into the analytical process. Rather than relying solely on external labels, procedural checks, or post hoc controls, forensic genomics can now leverage the data stream to carry information about sample provenance, workflow history, and authenticity.
This shift invites a broader question:
The Hidden Risk in High-Throughput Forensic Genomics
Modern forensic laboratories manage large volumes of evidence through complex, multi-step workflows. A single sample may pass through extraction, library preparation, sequencing, and multiple layers of bioinformatic analysis before a result is ever interpreted.
Even in highly disciplined laboratories, this complexity introduces risk.
Labels can be damaged or lost. Samples can be inadvertently swapped during handling. Trace amounts of DNA can carry over from one library to another. Some errors are immediately obvious. Others may not become apparent until long after results have been reported and some errors may never be detected at all.
For decades, forensic laboratories have relied on external safeguards to manage these risks. Tube labels, barcodes, and positive and negative controls play an essential role in QC. But these tools monitor the process around the sample. They do not interrogate the sample itself.
They tell us how the workflow was intended to operate. They do not, on their own, prove that it did for any specific sample.
As forensic workflows grow more complex and data-rich, this distinction becomes increasingly important. Most traditional QC systems live outside the DNA, while the evidence itself remains silent about its own history.
Embedding the Workflow Inside the Sample
In a recent peer-reviewed study, the Othram team introduced a new approach designed to address this gap: molecular etches. These molecules are synthetic DNA sequences that function as an internal molecular information management system for forensic genome sequencing.
Rather than labeling a tube or tracking a barcode in software, molecular etches are added directly to the DNA sample before sequencing. They move through the entire laboratory and sequencing workflow accompanying the evidence DNA.
By varying the sequence, each molecular etch, or combination of etches, can encode information such as the time of processing, the workflow or analysis type used, the order of processing, and laboratory context of each and every sample.
Because these etches are sequenced along with the human DNA, they create a molecular audit trail that is inseparable from the data.
If the wrong etches appear, or expected etches are missing, it is an immediate signal that something failed, independent of external labeling or procedural records.
Why This Internal Sample Coding Matters for Defensibility
When a result is challenged by a defense attorney, a prosecutor, a judge, or a reviewing expert, various questions arise. Traditionally, questions have focused on technology and statistical inferences, and these issues change over time as a science becomes generally accepted. Certain questions persist and are more specific about handling and workflow processes, such as:
Molecular etches directly address these questions by embedding provenance into the sequence data itself. From the raw data alone, it becomes possible to verify whether the sample followed the correct path through the laboratory.
This data tracking is especially important in forensic genome sequencing, where sensitivity is high and the consequences of error can be significant, and where errors detected only after interpretation may undermine downstream conclusions.
Sensitivity That Enables Sample Attribution, Not Just Detection
One of the most important outcomes of the validation work is not merely that molecular etches detect contamination. It is how sensitive they are, and what that sensitivity enables.
Molecular etches detect extremely minor sample-to-sample carryover, including trace transfers that would typically go unnoticed in routine forensic workflows. At higher sequence depths, even the smallest contamination events are visible.
Because molecular etches encode workflow- and case-specific information, they do not just signal that contamination occurred. They can attribute the source of the contamination or a sample swap. When an unexpected molecular etch appears, it is an immediate indication of contamination, signaling from which sample it originated, through which workflow context it passed, and whether the event is consistent with carryover, handling error, or library-level transfer.
While traditional controls may indicate that contamination exists in reagents, these controls rarely explain the source and may allow contamination to go undetected with sample specific issues. Molecular etches transform contamination from a vague concern into a traceable and explainable event.
Instead of asking whether something went wrong, laboratories can immediately identify issues well before issuing a report, asking what happened, when it happened, and which sample or samples were involved.
Auditability Is Not Optional
As forensic genomics becomes more powerful, expectations around auditability rise with it.
In the same way that modern financial systems rely on transaction logs and reconciliation, forensic identity systems should require verifiable molecular chains of custody, not just procedural ones.
Molecular etches support early detection of sample swaps, detection and attribution of carryover contamination, independent verification of sample authenticity, and scalable automated quality checks.
Importantly, these safeguards operate without altering or damaging the evidentiary DNA and without interfering with downstream analysis, extending rather than replacing existing forensic QA/QC frameworks.
Supporting the Identity Inference Pipeline
At Othram, we describe our work as an identity inference pipeline. This system takes biological evidence and transforms it into a defensible identity outcome. That pipeline spans evidence intake, DNA extraction and sequencing, profile generation, database search and relationship analysis, genealogical inference, confirmation, and reporting.
Molecular etches strengthen this pipeline by allowing every inference to rest on a trustworthy molecular foundation. These internal indicators do not replace existing QC. Rather, they extend QC into never-before-available sample-specific data.
As forensic laboratories adopt MPS, these QA/QC and auditability considerations apply whether DNA testing is performed in-house or outsourced. Agencies should evaluate not only analytical performance, but also how sample provenance and workflow integrity are embedded into the sequencing process itself.
Othram supports both approaches. For laboratories seeking to onboard forensic sequencing, Othram deploys complete, validated workflow solutions, including molecular etches, to enable a defensible transition to MPS-based methods. For agencies not yet ready to bring this work in-house, Othram operates the world’s first purpose-built forensic laboratory for forensic genetic genealogy. More forensic genetic genealogy cases have been solved using Othram technology than any other method.
Reach out to learn how we can help you upgrade your capabilities or help you with your next case.